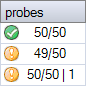
The probe counter in Coffalyser.Net provides a visual warning when an unexpected number of probes have been found in a sample. The probe counter has the following format:
number of probes found / number of probes expected | number of probes found that were not expected
The last component is only shown if there were unexpected probe signals.
It is important to inspect the data whenever the probe counter provides a warning, as an unexpected count can be caused by biological properties of the sample but also by experimental or data analysis issues.
Background
Probes for which a signal is expected
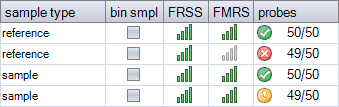
In many probemixes, all probes are expected to have a signal in healthy individuals. If a lower number of probes is found, this may be caused by a homozygous deletion, but also by e.g. an issue with peak identification (binning) or signals that are too low. The example below shows a probemix with 50 probes that are expected to produce a signal. Signals were found for all 50 probes in the first sample, but only for 49 probes in the second sample.
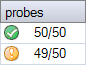
Probes for which a signal is not expected
Some probemixes have probes for which a signal is not expected in healthy individuals, such as mutation-specific probes. If a significant signal for a mutation-specific probe is found, this is unexpected and will result in a probe counter such as for the second sample in the screenshot below. The data should be inspected to determine if the sample might be positive for the mutation or if the result was caused by an artefact such as a nonspecific peak.
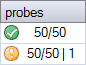
Differences between samples in the number of probes expected
The number of probes expected may differ between samples in the same experiment. If the probemix contains one or more probes for the Y chromosome, a signal for these probes is expected in male samples but not in female samples. In the example below there is one probe for the Y chromosome.

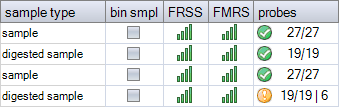
In MS-MLPA, the number of expected probes in digested and undigested samples may differ because some probes may be expected to produce no signal after digestion in healthy individuals. If the probemix has one or more digestion control probes, these are also expected to be absent after digestion. The example below is for a probemix in which eight probes are not expected to give a signal in the digested reaction. In the last digested sample, six of these nevertheless gave a signal. This may be due to the methylation status of the sample DNA, but could also be caused by e.g. incomplete digestion or nonspecific peaks.
Criteria for a probe to be counted in the probe counter
Probes are only counted as 'found' when their signal is:
- Above the minimum peak amplitude for probes (in RFU) defined for the electrophoresis device.
- Above 10% of the median signal of the reference probes.
Probes that are below 10% of the median signal of the reference probes but above the device's minimum signal may still be identified and present in the results, but will not be counted in the probe counter. This helps to more easily identify unexpectedly low signals using the probe counter.
In rare cases, slope correction may influence whether a probe signal crosses the abovementioned thresholds. As a result, the probe counter may change after the comparative analysis, when slope correction is performed. If this happens there were probably one or more probes with a signal just around the thresholds for inclusion in the probe counter.
Warnings for unexpected probe counts
An orange warning is displayed for an unexpected probe count. Test samples also get a subtraction to their FMRS score if a reference probe is missing, as this is very unusual. If reference samples are missing the signal of any expected probe, a red warning is displayed for the probe counter and the FMRS score gets a heavy penalty, because samples that miss an expected probe signal are not suitable as reference samples. In the example below, the second reference sample and the last test sample both miss a signal for a target probe, but this results in different warnings.