A bin set is a collection of bins, where a bin specifies the boundaries within which Coffalyser.Net searches for the signals of probes and fragments. By default, the boundaries of bins are 4 nucleotides apart to allow for some variation during electrophoresis.
For reliable analysis and result interpretation, it is important that the correct peaks are assigned to the correct MLPA probes and fragments. The lengths of these probes and fragments can differ slightly between samples within an experiment due to fluctuations in the electrophoresis run. For example, a fragment may be recognized as having a length of 140.25 nt in one sample, whereas it is recognized as having a length of 140.45 nt in another. The bin set is used to link such signals to the same probes. Because of the importance of a correct bin set, an incorrect bin set may lead to a lowered FMRS score.
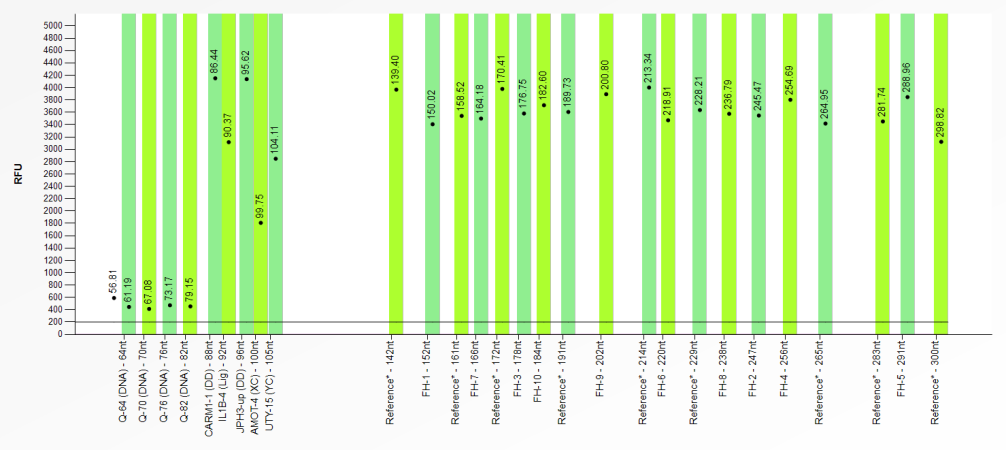
In the binning profile tab of the sample results in Coffalyser.Net, each bin is displayed as a vertical bar, and each recognised signal is displayed as a black dot with an accompanying size label. When a signal has been found near the centre of a bin, the bin is green. The image below is an example of a correct bin set where a signal has been detected for all bins. Therefore, all bins are green.
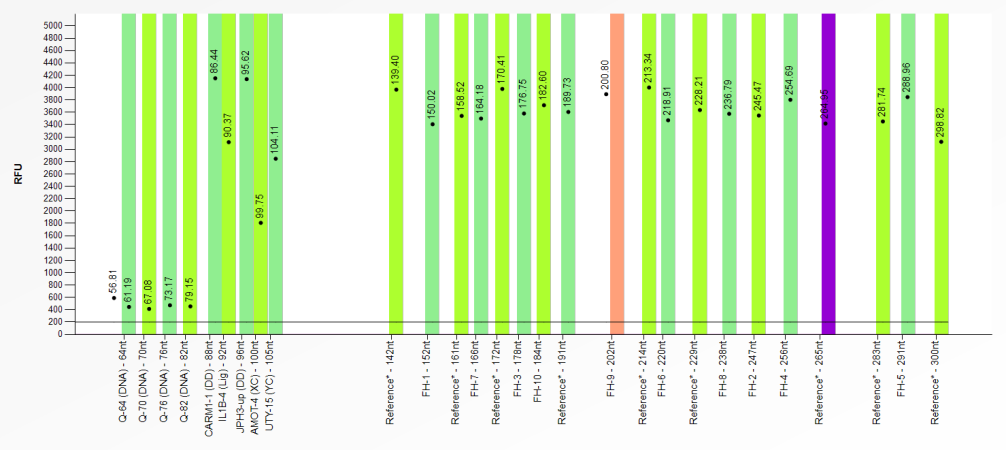
When no signal has been detected within a bin, it will be red. A purple bin indicates that a signal was found but that it is more than 0.7 nt away from the centre of the bin. The image below is an example of an incorrect bin set displaying both situations.
To check if the bin set is correct and all detected MLPA probes and fragments fall within a bin, follow the steps below:
- Right-click on a sample in the fragment analysis tab of an experiment in Coffalyser.Net.
- Select Open.
- Navigate to the binning profile tab.
- If desired, check other samples by selecting them from the list on the left side of the window.
Alternatively, the bin set can be checked in the manual bin set editor. Here the bin set can directly be adjusted if necessary:
- Right-click on a sample in the fragment analysis tab of an experiment in Coffalyser.Net.
- Select Edit Manual Bin Set Default Channel.
- If desired, check other samples by selecting them from the list on the left side of the window.
Instructions on how to correct a bin set by creating a manual bin set can be found in this article.