The digitalMLPA™ technique has been designed in such a way that library preparation can be done quickly and easily without the need for purification, quantification or assessment before sequencing. However, we understand that some customers still like to check that there was successful amplification of the ligated digitalMLPA probes. Analysis using traditional NGS library assessment methods such as a TapeStation can be used, but with some additional guidelines.
Using a TapeStation
If it is desired to visualise a digitalMLPA library before loading on an Illumina sequencer this can be done using an Agilent TapeStation. The TapeStation is a non-denaturing electrophoresis system that is commonly used for the assessment of NGS libraries. This system must be used with caution, as sample DNA and carrier DNA will be visible along with digitalMLPA amplicons. The majority of DNA present (80–90%) will be digitalMLPA amplicons visible in the 190–220 bp range.
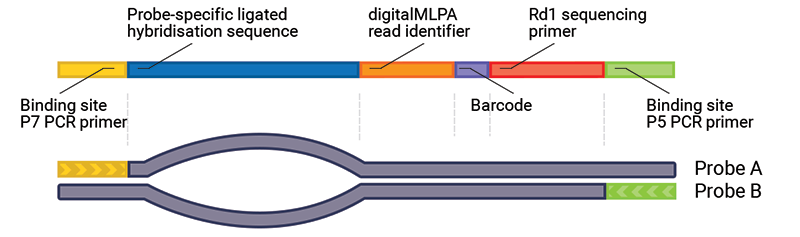
After the digitalMLPA reaction, all amplicons are single-stranded or part of heterodimer complexes. For correct visualisation, the TapeStation sample should first be incubated for ≥3 hours at 60ºC. This allows for all amplicons to become double stranded (forward and reverse strand of the same probe), enabling the correct fragment visualisation at ~200 bp.

Artifacts without incubation at 60°C
If the additional incubation period at 60ºC is not performed, larger fragments than expected may be visible. This is due to the formation of mixed hybrids: products formed from the forward strand of one probe and the reverse strand of a different probe. Mixed hybrids are formed quickly after the PCR due to the homology between the sequences at both ends of all probes (e.g. primer sequences, digitalMLPA read identifier). These mixed hybrids still have the same fragment length but migrate at a much slower rate due to the non-hybridising “bubble” between the double stranded ends. It is also possible that more complex hybrid structures form, further reducing fragment mobility.