When reaction failure is suspected, it is possible to visualise digitalMLPA fragments by capillary electrophoresis to verify that digitalMLPA PCR products are present. This check can be done on an ABI capillary electrophoresis device before sequencing on an Illumina-based sequencing platform. Analysis by capillary electrophoresis is intended for troubleshooting purposes only.
Procedure
If sequencing on an Illumina sequencer fails, equipment failure during a digitalMLPA run occurred, or reaction failure is suspected due to another reason, it is possible to visualise digitalMLPA fragments on an ABI capillary electrophoresis device. The forward P7 PCR primer used in the digitalMLPA PCR reaction is labelled with FAM fluorescent dye, enabling PCR product visualisation on an Applied Biosystems capillary sequencer. The FAM label does not interfere with Illumina sequencing.
Electrophoresis specifications
PCR products from a digitalMLPA reaction should be diluted in ultrapure water before use in an injection mixture. The optimal dilution factor may require optimization, but we suggest starting with a dilution factor of 10. For example, take 1 µl of digitalMLPA PCR products and mix with 9 µl of ultrapure water.
Instrument |
Primer Dye |
Capillaries |
Injection Mixture |
|---|---|---|---|
ABI-Prism 3100 (Avant) ABI-3130 (xL) ABI-3500c (xL) ABI-3730 (xL) |
FAM |
36, 50 cm |
|
ABI-SeqStudio |
FAM |
28 cm |
|
The volume of PCR product added should never exceed 10% of the total injection mixture. Only use diluted digitalMLPA PCR products.
Reduce volume of size standard if needed.
For the ABI-3500: set run voltage to 15 kV and ensure sufficient run time.
Briefly heating the injection mixture before capillary electrophoresis is recommended.
Expected digitalMLPA peak pattern
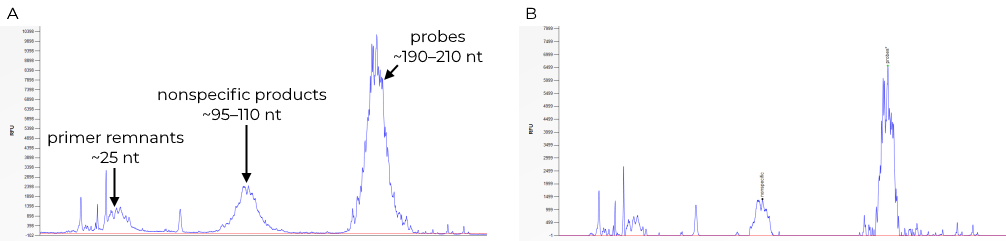
Most digitalMLPA probes generate amplicons between 190 and 210 nt. These amplicons will result in a broad, usually off-scale, peak between these fragment sizes (see Figure 1). This broad 190–210 nt peak should be the dominant peak visible in your peak pattern. Individual fragments are not visible. Q-fragment peaks, used for DNA quantity estimation, may be visible at around 145 and 150 nt when low amounts of sample DNA are used. Primer remnants, modified primers, and primer breakdown products may be visible in the electropherogram around 25 nt. An additional broad peak between 95 and 110 nt will likely also be visible in your peak pattern. This and other distinct visible peaks not specifically mentioned are the result of non-specific amplification products that will not affect your digitalMLPA reaction.
There are two options to view digitalMLPA troubleshooting results from a CE device.
The electropherogram can be viewed in Coffalyser.Net, our free analysis software for MLPA. The raw data can be evaluated in the raw data tab of the sample results explorer. For optimal results, download the digitalMLPA troubleshooting sheet for Coffalyser.Net. This sheet can help you identify the probes peak at 190–210 nt and the nonspecific peak between 95 and 110 nt (Figure 1B). To use the sheet, import it in Coffalyser.Net, set the analysis method to population, set the probe recognition method to manual during fragment analysis, and view the pattern in the genomic profile sheet of the sample results explorer. Adjust the zoom level as needed.
The electropherogram can be viewed on your capillary electrophoresis device.
digitalMLPA reaction failure peak patterns
When reaction failure has occurred the expected large peak between 190 and 210 nt will be absent or severely diminished. A sample where a digitalMLPA reaction with the same digitalMLPA probemix was successful can be used for comparison. NOTE: Peak patterns are not expected to be identical, some variation will be present.
In a failed reaction, the Q-fragment peaks at 145 and 150 nt are usually still visible. If this is not the case, this may be explained by e.g. the omission of probemix or polymerase from the reaction, or by impurities in the DNA sample that fully inhibited polymerase activity.
In case of reaction failure check your reagents, sample quality, and equipment before repeating your digitalMLPA reaction. Ensure all steps outlined in the digitalMLPA NXtec Protocol are followed exactly as written.
Precautions
Baseline correction by ABI analysis software will alter the peak pattern image in relation to the one displayed in our guide (Figure 1). Any image where a large peak is visible in the 190–210 nt range indicates the reaction was successful.
The cause of digitalMLPA reaction failure cannot fully be determined by examining the peak pattern generated by capillary electrophoresis. Sequencing of suspected failed reactions may also give insight into the cause of the reaction failure as every digitalMLPA probemix includes control probes for enzyme activity and DNA quality.
Additional information
Non-specific peaks smaller than 120 nt may generate sequencing clusters but most will not generate reads.
Non-specific peaks larger than 120 nt may generate reads but almost all will be removed during analysis by Coffalyser digitalMLPA.
For additional digitalMLPA troubleshooting advice please see the digitalMLPA NXtec Protocol or contact us.