To prevent sample depurination, it is essential that DNA samples contain sufficient buffering capacity during the initial denaturation step of the MLPA and digitalMLPA protocols. The presence of at least 5 mM Tris-HCl pH 8.0–8.5 in a sample is sufficient to prevent depurination.
In an MLPA reaction, if it is not known what the sample DNA is dissolved in, we recommend that 1 µl of 50 mM Tris-HCl pH 8.5 is added to 4 µl of sample DNA.
Background
During MLPA and digitalMLPA reactions depurination can occur during the initial denaturation step if the sample DNA is in a solution with insufficient buffering capacity or a low pH (e.g. water). Certain sample treatments (e.g. prolonged heating, formalin fixation/paraffin embedding) can also result in depurination of the sample DNA, even with sufficient buffering capacity. Problems are more likely to arise when these factors are combined.
Depurination of DNA occurs when a purine base (adenine (A) or guanine (G)) is removed from the DNA deoxyribose backbone via hydrolysis. This results in a deoxyribose lacking a purine base, which is known as an abasic or apurinic site.
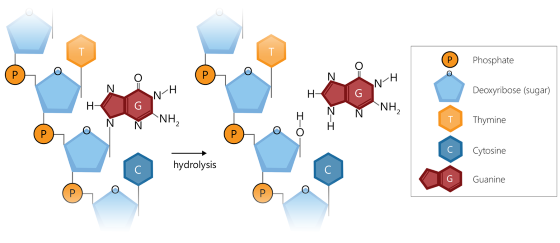
Depurination at the ligation site of a probe, especially if it has a large number of pyrimidines (cytosine (C), or thymine (T)), will result in inefficient probe hybridization and subsequent limited or failed probe ligation, as these pair with the purines (G, A) that are lost during depurination. As a consequence, the probe signal will be reduced or even absent. Loss of probe hybridization/ligation also leads to variability in the obtained probe signals.
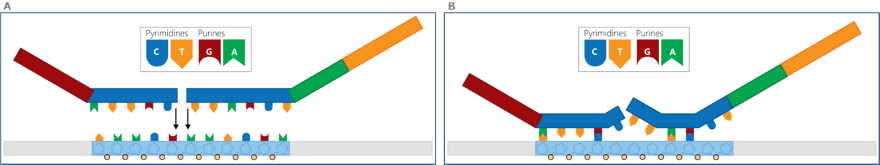
The figure below shows a schematic representation of a probe with high pyrimidine content around the ligation site (i.e. purines present in the target DNA). In panel A, the probe hybridises successfully to the sample DNA, which in this case is unaffected by depurination. In panel B, the same probe is hybridised to sample DNA that is heavily affected by depurination: the guanines and adenines of the target on the sample DNA have all been lost. The result is a very poor probe hybridization, leading to an inefficient or failed ligation of the probe oligonucleotides, ultimately causing a reduced or even absent probe signal.