MLPA (Multiplex Ligation-dependent Probe Amplification) is a method that detects aberrant copy numbers in up to 60 specific nucleic acid sequences by performing one simple PCR reaction, using a single PCR primer pair. MLPA reactions require only 50 ng of human genomic DNA. Applications include: the detection of exon deletions/duplications in e.g. the human BRCA1, MSH2 and MLH1 genes, detection of trisomies such as Down syndrome, characterisation of chromosomal aberrations in cell lines and tumour samples, SNV detection, and DNA methylation analysis.
Conventional multiplex PCR uses one pair of primers for each fragment, resulting in a reaction that contains a large number of different primer sets. The primer efficiency of different primer pairs can vary, thus making it difficult to use conventional multiplex PCR for relative quantification of target sequences. Furthermore, small differences in reaction conditions can result in large differences in the obtained results as each primer pair may react slightly differently to the change.
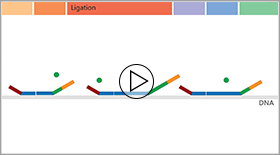
In contrast, all fragments in an MLPA reaction are amplified using the same PCR primer pair, making the reaction extremely robust. The trick of MLPA is that it is not the sample DNA that is amplified, but the MLPA probes that are added to the sample. Each MLPA probe consists of two paired oligonucleotides, each one containing one of the PCR primer sequences plus a sequence complementary to the DNA target sequence. These two probe oligonucleotides hybridise to immediately adjacent target sites. Only when the two probe oligonucleotides are hybridised to their target can they be ligated into a single probe, containing both the forward and reverse primer sequence. These ligated probes are amplified exponentially during the PCR reaction, while the individual non-ligated probe oligonucleotides are not. The number of probe ligation products of one probe therefore depends on the number of target sequences in the sample.
Following the PCR, the resulting amplification products are separated by capillary electrophoresis. An MLPA probe set is designed so that the length of each of its amplification products is unique. The length increases in a stepwise-fashion, with the total size range lying between 120-500 nucleotides. This size range provides optimal fragment separation and low background on sequence type gels.
The only equipment needed to perform MLPA is a capillary electrophoresis device, and a thermocycler. MLPA reactions can be performed as a one tube reaction, as non-ligated oligonucleotides do not need to be removed.
Click on the image below to watch an animation that explains the principle behind MLPA in more detail, or continue reading about the advantages and limitations of MLPA.